Knowledge
Knowledge
Resources
The Resources section is a curated collection of interpreted materials, including research articles, publications, and summaries of presentations, that support understanding and discussion of genomic surveillance and related topics in public health. These resources provide context, insights, and learning opportunities for public health professionals.
Genomic Surveillance
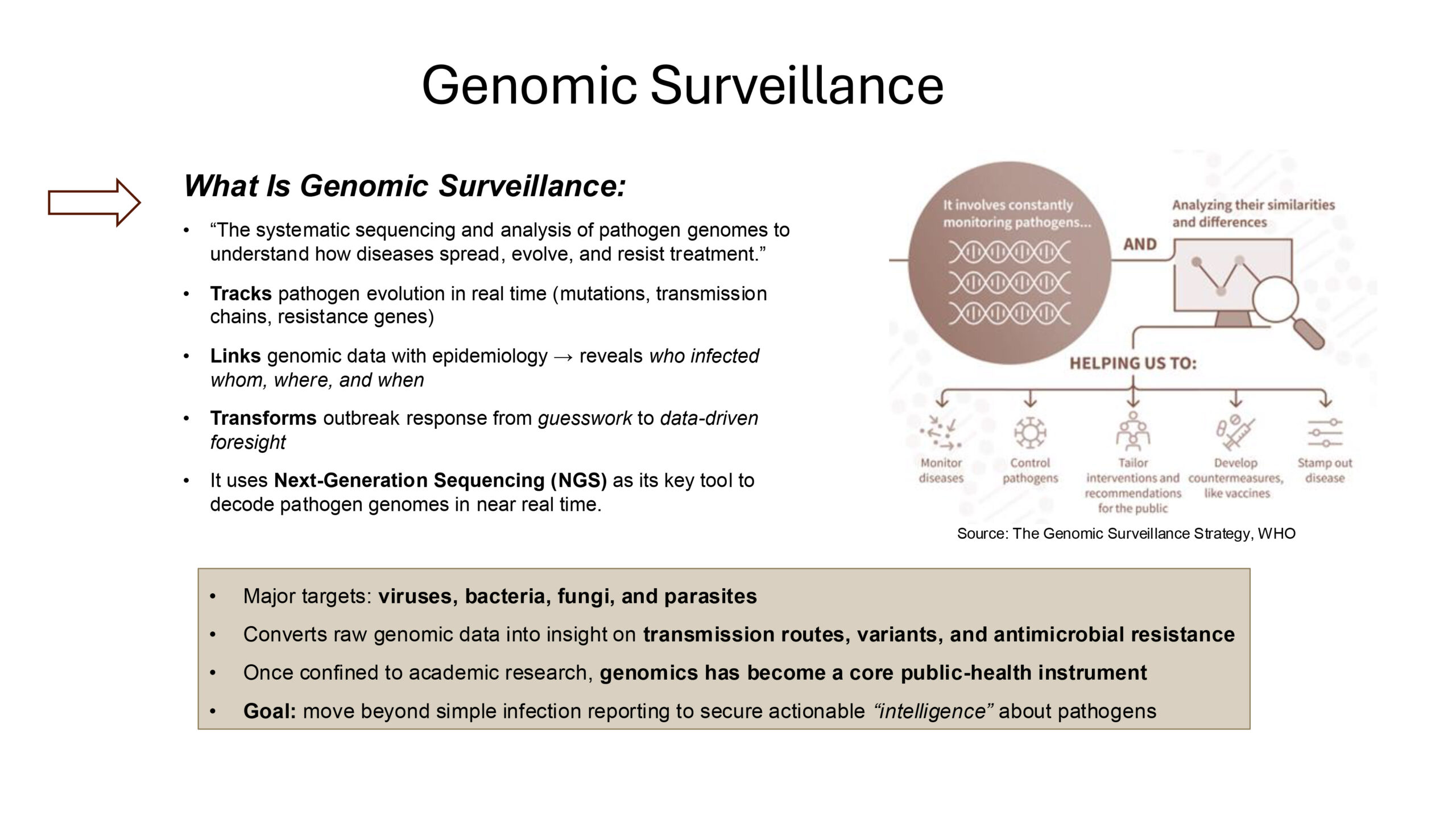
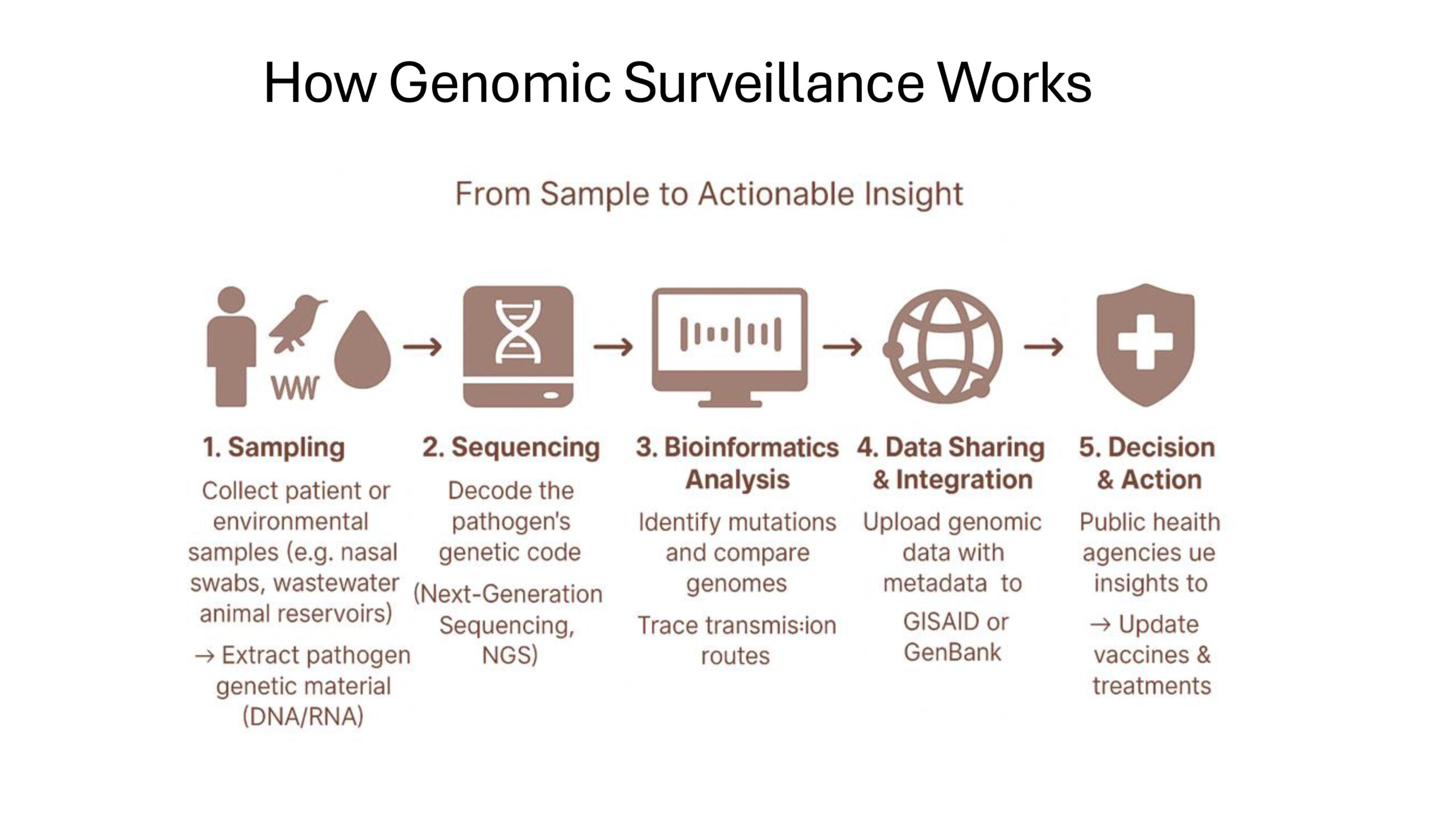
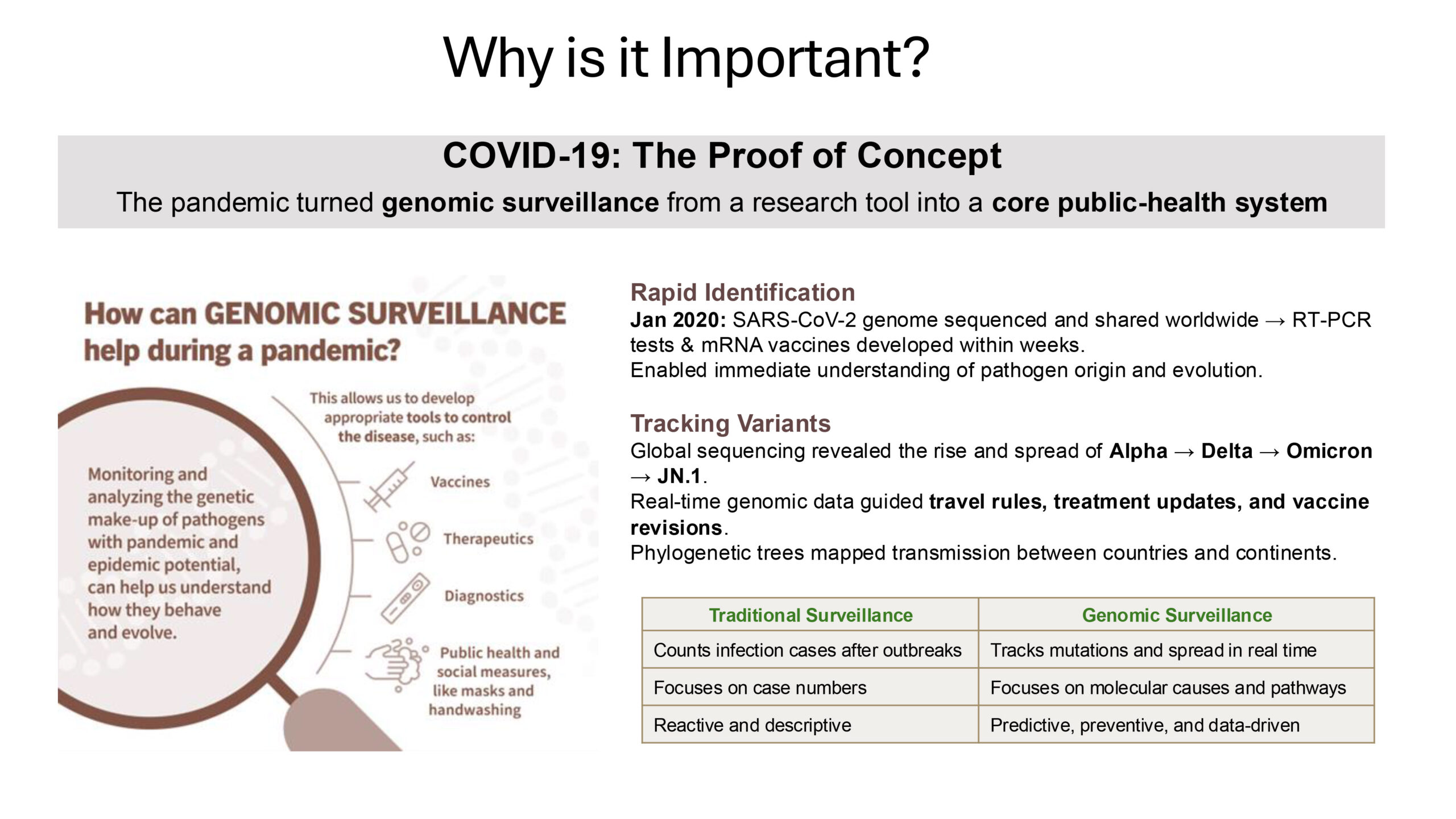
Source: The Genomic Surveillance Strategy, WHO
New Literature
1. Global landscape of SARS-CoV-2 genomic surveillance and data sharing
This study provides a comprehensive assessment of global SARS-CoV-2 genomic surveillance and data-sharing practices across 118 countries during the COVID-19 pandemic. By analyzing millions of viral genomes, it reveals substantial inequities in sequencing capacity, turnaround time, and metadata completeness between high-income and low- and middle-income countries. The findings show that effective variant detection depends not only on sequencing volume but also on representative sampling and standardized data sharing. These insights underscore the need to embed genomic surveillance as a routine, decentralized component of public health systems rather than relying on ad-hoc emergency sequencing efforts.
2. Genomic epidemiology in public health surveillance
Genomic epidemiology has become a core component of public health surveillance by integrating pathogen genomic data with epidemiological information. This article explains how genomic data are used to track transmission dynamics, detect emerging variants, and support routine surveillance and outbreak investigation. It provides a clear interpretation of how genomic surveillance has evolved from a research activity into an applied public health function.
3. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus
This study shows how genomic surveillance enabled rapid characterization of the 2022 monkeypox outbreak, revealing accelerated viral evolution and sustained human-to-human transmission. It highlights the value of genomic surveillance for tracking non-respiratory pathogens and identifying evolutionary changes relevant to public health response.
4. The pandemic gap of respiratory viruses during the COVID-19 pandemic
Using long-term molecular surveillance data, this article describes how the COVID-19 pandemic reshaped global respiratory virus ecology. It introduces the concept of the “pandemic gap,” demonstrating how genomic and molecular surveillance revealed prolonged suppression and later re-emergence of common respiratory viruses, with implications for population immunity and preparedness.
5. Monitoring antimicrobial resistance trends from global genomics data: amr.watch
This study demonstrates how large-scale genomic datasets can be used to monitor global trends in antimicrobial resistance. It highlights the public health value of genomic surveillance for tracking resistance genes and lineages, while also identifying limitations caused by missing or incomplete metadata.
6. Genomic Surveillance for Viruses of Public Health Importance in Low- and Middle-Income Countries: Opportunities and Challenges
This article examines the opportunities and structural challenges associated with implementing genomic surveillance in low- and middle-income countries. It discusses issues related to sequencing capacity, workforce development, data sharing, and sustainability, providing a grounded interpretation of how genomic surveillance can be adapted to resource-limited public health systems.
Link: https://www.tandfonline.com/doi/full/10.1080/14737159.2025.2543750
doi: https://doi.org/10.1080/14737159.2025.2543750">https://doi.org/10.1080/14737159.2025.2543750
doi: https://doi.org/10.1080/14737159.2025.2543750">https://doi.org/10.1080/14737159.2025.2543750
7. Implementation of genomic surveillance of SARS-CoV-2 in the Caribbean: Lessons learned for sustainability in resource-limited settings
This study documents the implementation of genomic surveillance for SARS-CoV-2 in Caribbean countries and analyzes lessons learned regarding sustainability, regional collaboration, and capacity building. It provides practical insights into how genomic surveillance systems can be maintained beyond emergency response in small and resource-limited public health settings.



